Help
|
Input The user has to supply single or multiple peptide sequence to run the iAMPpred. The sequences must be supplied in FASTA format. Each sequence should contain at least 10 amino acids. Moreover, each sequence should contain only standard amino acid residues in a single letter code format i.e., A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y. An example dataset having 5 peptide sequences is given below. >AP00036 DFASCHTNGGICLPNRCPGHMIQIGICFRPRVKCCRSW >AP00037 VRNHVTCRINRGFCVPIRCPGRTRQIGTCFGPRIKCCRSW >AP00038 QGVRNHVTCRINRGFCVPIRCPGRTRQIGTCFGPRIKCCRSW >AP00039 QRVRNPQSCRWNMGVCIPFLCRVGMRQIGTCFGPRVPCCRR >AP00040 QVVRNPQSCRWNMGVCIPISCPGNMRQIGTCFGPRVPCCRRW OutputThe out put is provided in a tabular format as given below. The table shows the probability with which the peptide sequences are predicted as antibacterial, antiviral and antifungal. If the probabilty is >0.5, then the test peptide sequence is said to be predicted in positive class and other wise negative class. Moreover, if a instance is predicted in more than one positive classes, then the liklihood of that instance as an AMP will be higher and vice versa. 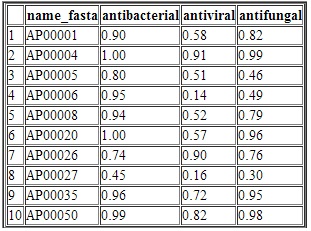
|