Input for prediction with miRNA dataset: User has to supply the miRNA sequences in FASTA format. Each sequence should contain ony the standard nucleotides i.e., A, U, G and C. An example of two sequences is given below.
>zma-miR169e MIMAT0001736
UAGCCAAGGAGACUGCCUACG
>ath-miR399b MIMAT0000952
UGCCAAAGGAGAGUUGCCCUG
Input for prediction with Pre-miRNA dataset: User has to supply the Pre-miRNA sequences in FASTA format. Each sequence should contain ony the standard nucleotides i.e., A, U, G and C. An example of two sequences is given below.
>zma-MIR169e MI0001834 Zea mays miR169e stem-loop
GCAAUAGGGGCCACUCAGGCUAGCCAAGGAGACUGCCUACGAACCAACACAAAGGUCCACAAUUCUGAUCCUUUGUAAAC
AAAGGACAUAGGCAAGUCAUCCUUGGCUAUCAGAGGCAGGCCCUUAUU
>ath-MIR399b MI0001021 Arabidopsis thaliana miR399b stem-loop
UCACUAGUUUUAGGGCGCCUCUCCAUUGGCAGGUCCUUUACUUCCAAAUAUACACAUACAUAUAUGAAUAUCGAAAAUUU
CCGAUGAUCGAUUUAUAAAUGACCUGCCAAAGGAGAGUUGCCCUGAAACUGGUUC
Input for prediction with Pre-miRNA+miRNA dataset: User has to supply the both Pre-miRNA and miRNA sequences in FASTA format, where the Pre-miRNA and miRNA sequences are corresponding to each other. Each sequence should contain ony the standard nucleotides i.e., A, U, G and C.
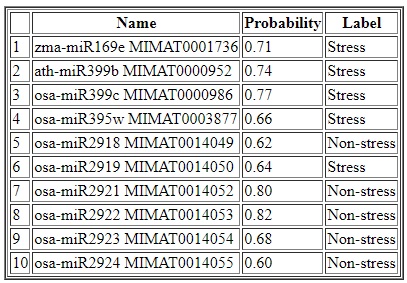
Output for prediction: The result table shows the probabilities with which each sequence is predicted as stress-responsive or non-stress-responsive.The table below shows the predited label as well as the probability with each sequence is predicted. For instance, the first sequence is predcited as stress-responsive with probability 071. Similar inference can be amde for other sequences as well.