Help
Input
The input sequences must be supplied in FASTA format. Each sequence should contains only standard amino acid residues in a single letter code format i.e., A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y. An example dataset having 3 protein sequences is given below.
>Seq_1 MMINYWNPIEEIDTVRRQLDHLFEDAIDTGKSSNYPSWAPAVELWDTGDALILKAFLPGVNADSLDIQATRESISIA GERHPEDLQEGTKRLFSDINYGHFRRASKLPVAIQNTKVEASFEQGLLTLRLPKVEEEQNKVVKVNLLNKGETSQAA LEASHNS >Seq_2 MMSIVLRDPFRSFERMYPLGWEPFQELESWRREMDRMFGRLMPISKSDGEKSLTFMPSAEMDETDQEIHLKFELPGL DAKDLDIEVTKDAVYIRGERKTEIEAESEGTVRSEFHYGKFERVIPMPSPIKTDNVQAEYNNGVLSLTLSKSEEDMK KSVKVEVS >Seq_3 MVDSQDHYSVLQVSPQATPADIKTAFRRLVRQYHPDLNPNNPRAAEAFQKICTAYEVLSNRDRRVIYDHNDGPRSPF VEEPGSVVLQDAQQYFMQGVQKANRRNYATAIADFTQAIHLDKNYLEAYMGRCQARFALGHNREVLQDCDHILSIQP QSAQAFFFRGRSCYRLGHLDLSIESYSQAIALEKDYAQAFYHRGIAYIKVKERLAIRDLQAAASLFRQQGHIGHYQR AIATLKDLNRKPANILLNLPRNGATLATTALTTLPRLLVNPSGAALQVWQSQLPYQSIGISLICAGLAMGCVVGGAH LFAPKPWTVSDSQVMLLSLVAFSSLVLAGSLIRRMVGKGGSKGGSWSGDLLVAGSAMLPVGVWAFLSGLAMQLGQIE LIALSLFASSFTILGLYSGYTKIGQIAEPIAALAVPTIVMISYGVTALLYRALMLQLV
Output
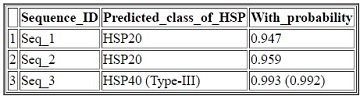
The output (results) are displayed in a tabular format with four columns. The first column represents the serial number, second column represents the sequence identifier, third column represents the types of predicted HSP (with subtype of DnaJ if predicted as HSP40) or non-HSP and the fourth column represents the probabilities with which they are predicted in the respective classes. For example, the second sequence is predicted as HSP20 with probability 0.959, whereas the third sequence is predicted as HSP40 with probability 0.993 and again this HSP40 is predicted as Type-III DnaJ protein with probability 0.992.