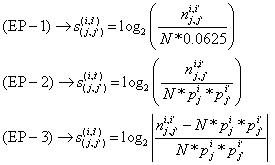Di-nucleotide association score
Following are the adjacent di-nucleotide associations scores computed under the three types of encoding procedures.
- EP-1: Association between nucleotides of two adjacent positions was expressed in terms of the ratio of observed frequency to the frequency due to random occurrence. The denominator of the ratio is expressed as N times of 0.0625, where N is the number of aligned sequences and 0.0625 is the probability of occurrence of di-nucleotide together at random (0.25*0.25). The observed frequency of occurrence for a pair of nucleotides at adjacent positions was measured from all the total set of splice site motifs, by counting the occurrence of such pairs of nucleotides. The score was measured in terms of logarithm scale with base 2 to store the information in bit and to bring the scores to normal.
- EP-2: Association between nucleotides of two adjacent positions was expressed in terms of the ratio of observed frequency to the expected frequency, where expected frequency is computed from PWM under the assumption of independence between positions.
- EP-3: Adjacent di-nucleotide association was expressed in terms of the relative deviation of observed frequency from the expected frequency computed from PWM.
The mathematical expressions for the proposed di-nucleotide association scores between two adjacent nucleotides is explained as follow;
Let 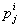 is the probability of occurrence of is the probability of occurrence of 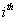 nucleotide at nucleotide at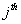 position. position. 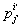 is the probability of occurrence of is the probability of occurrence of 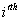 nucleotide at nucleotide at 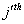 position. position. 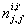 is the frequency of occurrence of is the frequency of occurrence of 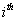 and and 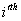 nucleotide together at nucleotide together at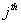 and and  positions respectively. Then the three different di-nucleotide association scores between positions respectively. Then the three different di-nucleotide association scores between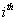 and and 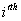 nucleotides occurring at nucleotides occurring at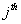 and and 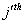 position under EP-1, EP-2, EP-3 was calculated using following formula position under EP-1, EP-2, EP-3 was calculated using following formula
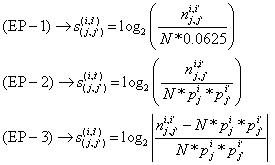
respectively, where 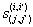 is the association score, N is the total number of sequence motif in the data set and is the association score, N is the total number of sequence motif in the data set and 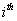 , ,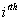 Є{A,T, G, C} and Є{A,T, G, C} and 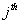 = 1, 2, …, 101 ; = 1, 2, …, 101 ; 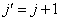 . A pseudo count of 0.001 was added to avoid logarithms of zero in the frequencies. . A pseudo count of 0.001 was added to avoid logarithms of zero in the frequencies.
Encoding ofsplice sitemotifs
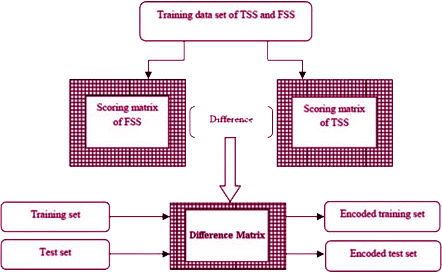
Since the sequence length of eachsplice site motif is of 102 nucleotides, 101 (=102-1) combinations of two adjacent positions are possible. Again, in each combination, 16 possible pairs of nucleotides (AA, AT,…,CG, CC) are possible. Thus, scoring matrices, each of order 16×101, were constructed using di-nucleotide association scores under all the three procedures. Then eachsplice site motif was encoded into a vector of scores of length 101 from the scoring matrix. So for N each sequence one will get a numeric data set of N observations and 101 variables (features). The diagrammatic representation of encoding splice site motif is given below
|