PreDOSS: A webserver for prediction of donor splice sites in eukaryotes

Input & Output for PreDOSS
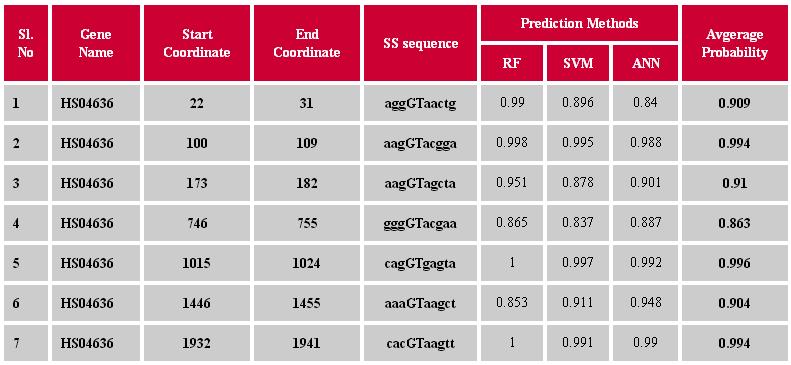
Input The input sequence data should be provided in single or multiple sequences in FASTA format. An example of input sequence file is Output The result provided in the table below is obtained after executing all the three machine learning techniques.The generated output file contains the probabilities with which the putative splice sites are predicted as true by different machine learning approaches. Higher is the probability more is the predictive strength. The last column contains the probabilities, averaged over prediction methods used. Here, the results of the putative splice sites with average probability >0.65 are displayed. However, the recommended threshold value and methods are explained below under "Prediction using threshold value".
Prediction using threshold value Among all machine learning techniques, though ANN achieved higher accuracy it is recommended to use all the three classifiers for the prediction of donor splice sites and take the decision on majority basis. In other words, if a putative splice site is predicted as true by any of the two methods then it can be declared as true splice site.
|
Team: Prabina Kumar Meher, Tanmaya Kumar Sahu, A. R. Rao and S. D. Wahi; Contact: meherprabin@yahoo.com